Probabilistic clustering of sequences: inferring new bacterial regulons by comparative genomics.
Posted in Publications, publishedNo Comment
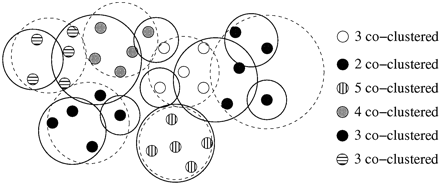
van Nimwegen E., Zavolan M., Rajewsky N., Siggia E.D. Probabilistic clustering of sequences: inferring new bacterial regulons by comparative genomics. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(11), 7323-7328.
Abstract
Genome-wide comparisons between enteric bacteria yield large sets of conserved putative regulatory sites on a gene-by-gene basis that need to be clustered into regulons. Using the assumption that regulatory sites can be represented as samples from weight matrices (WMs), we derive a unique probability distribution for assignments of sites into clusters. Our algorithm, “PROCSE” (probabilistic clustering of sequences), uses Monte Carlo sampling of this distribution to partition and align thousands of short DNA sequences into clusters. The algorithm internally determines the number of clusters from the data and assigns significance to the resulting clusters. We place theoretical limits on the ability of any algorithm to correctly cluster sequences drawn from WMs when these WMs are unknown. Our analysis suggests that the set of all putative sites for a single genome (e.g., Escherichia coli) is largely inadequate for clustering. When sites from different genomes are combined and all the homologous sites from the various species are used as a block, clustering becomes feasible. We predict 50–100 new regulons as well as many new members of existing regulons, potentially doubling the number of known regulatory sites in E. coli.
Short URL: https://tinyurl.com/y5r97w7z








Comments and Reactions
No comments yet. Why don't you write one?